Project information
- Category: Machine Learning, Multi-class Classification
- Source Data: Download
Project Details
Data Description
Iris flower classification is a popular dataset in machine learning project. The dataset contains three classes of flowers: Versicolor, Virginica, Setosa and each class has 4 features like “Sepal length”, “Sepal width”, “Petal length” and “ Petal width”. The goal of the iris flower classification is to train the SVM mode and predict flowers based on their features.
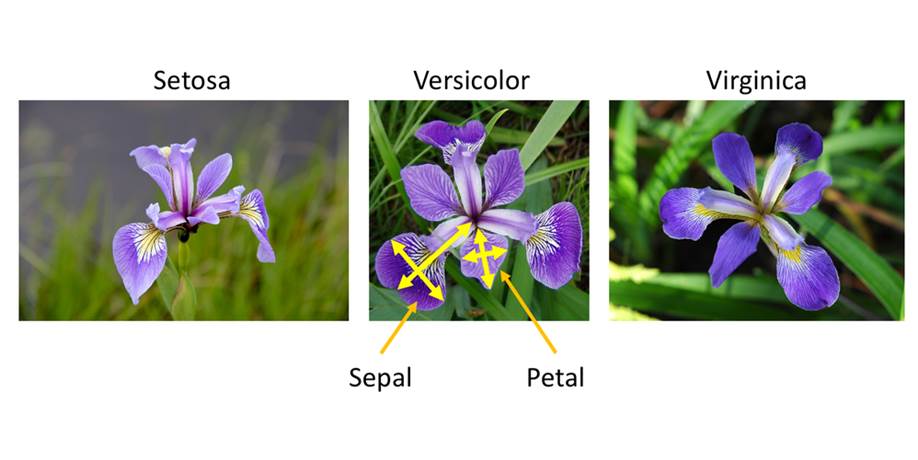
As the dataset have labeled the variable, it is supervised machine learning. Supervised machine learning are types of machine learning that are trained on well-labeled training data. Labeled data means that training data is already tagged with correct output.
In this project, I will solve the problem using the algorithm: Support vector machine as it is a supervised machine learning algorithm which analyzes data for classification and regression. Also, SVMSs are one of the most robust classification methods.
How SVM works?
SVM approximates a line (hyperplane) separating data between two classes.
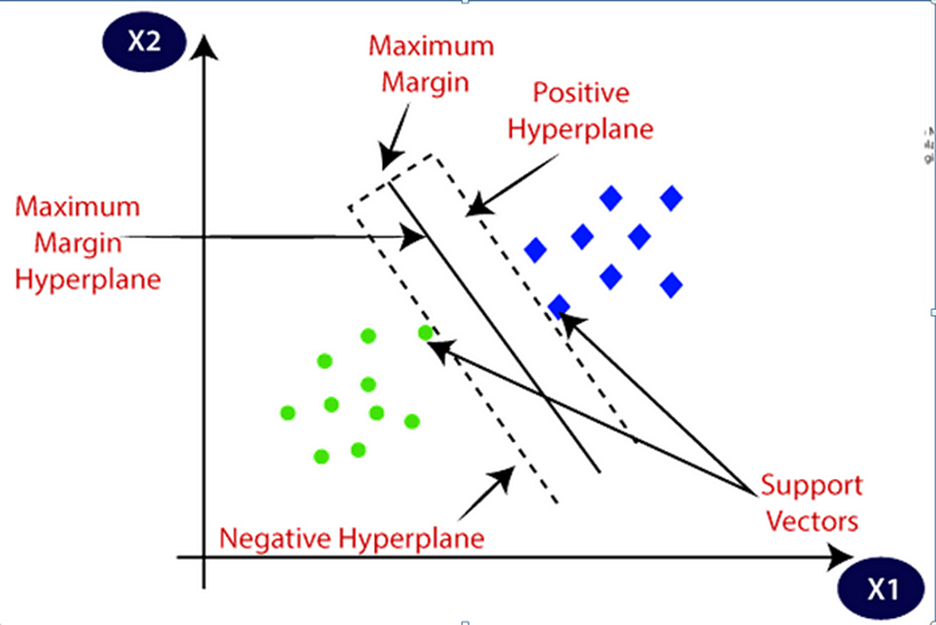
SVM algorithm finds the points closest to the line form two classes. These points are known as support vectors. Then, it computes the distance between the line and support vectors. The distance is called margin. The main purpose is to maximize the margin. The hyperplane which has maximum margin is known as the optimal hyperplane.
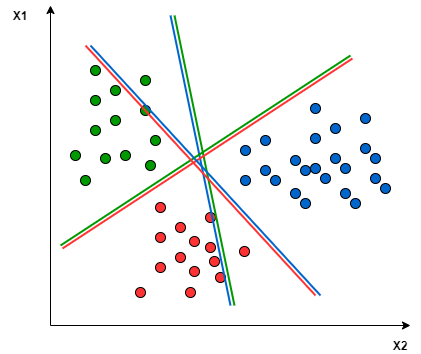
It mainly supports binary classification natively. For multiclass classification, it separates the data for binary classification and uses the same method repeatedly to break down multi-classification problems into multiple binary classification problems.
Python Packages:
- Numpy
- Matplotlib
- Seaborn
- Pandas
- Scikit-learn
Roadmap:
- Load the data
- Analyze and visualize the dataset
- Split a dataset into training and testing datasets
- Train the model
- Model Evaluation
- Testing the model
Step 1:
Import packages:
#import libaries import numpy as np import matplotlib.pyplot as plt import seaborn as sns import pandas as pd from sklearn.model_selection import train_test_split from sklearn.svm import SVC from sklearn.metrics import accuracy_score from sklearn.metrics import classification_report
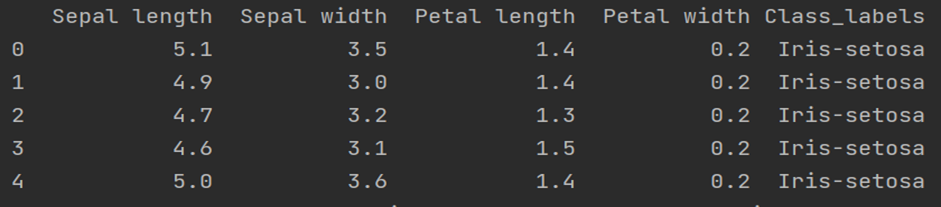
csv_url = 'url' columns = ['Sepal length', 'Sepal width', 'Petal length', 'Petal width', 'Class_labels'] # Load the data df = pd.read_csv(csv_url, names=columns) # display first 5 rows data print(df.head())
Step2: Analyze and visualize the dataset.
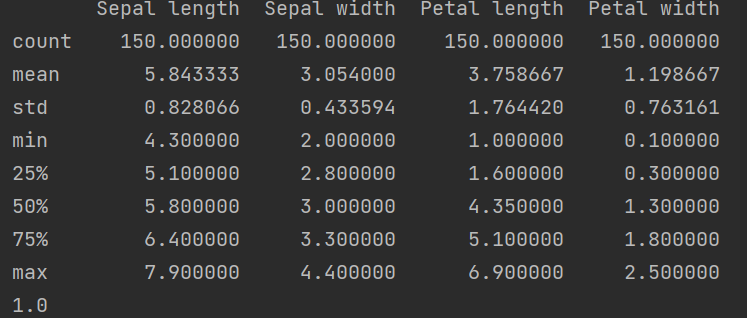
# display base statistical analysis regarding the data print(df.describe())
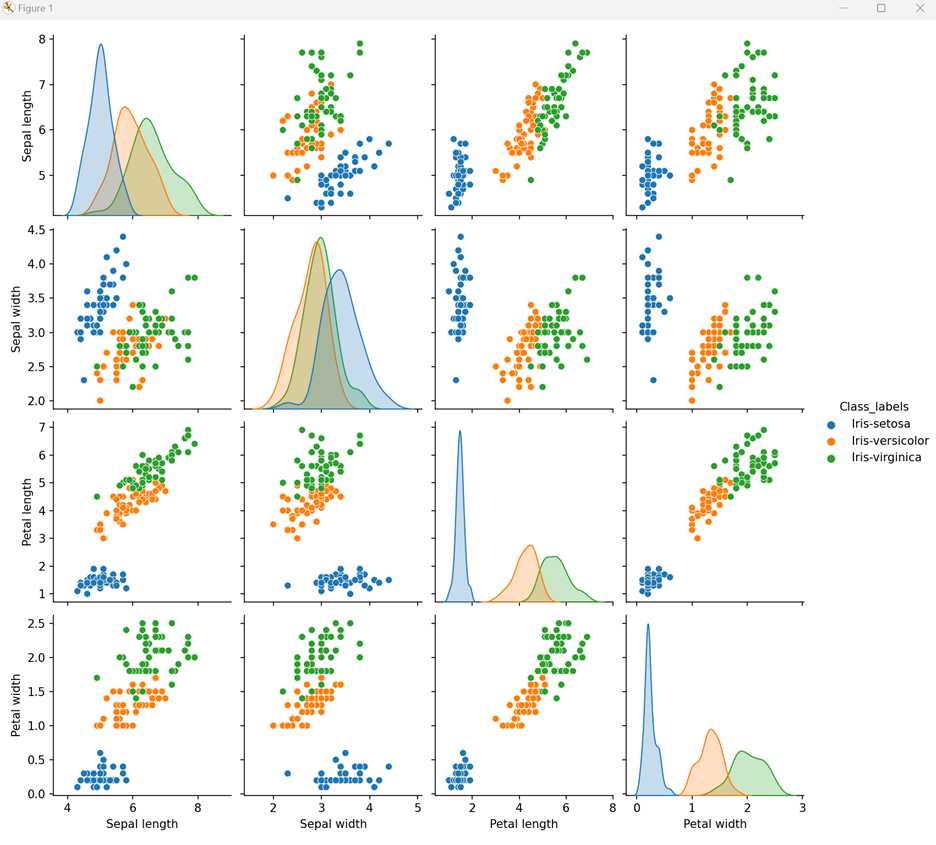
# visualize the dataset by labeled variable sns.pairplot(df, hue='Class_labels') plt.show()
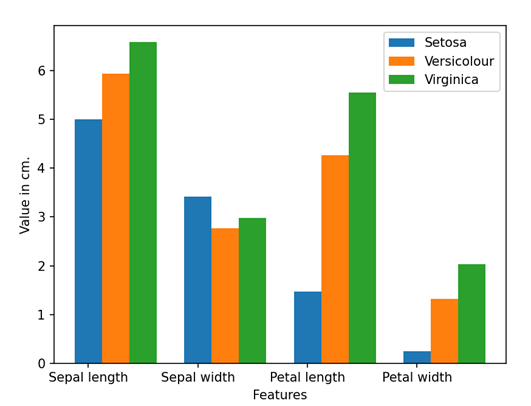
# Separate features and target data = df.values X = data[:,0:4] Y = data[:,4]
# Calculate average of each features for all classes
Y_Data = np.array([np.average(X[:, i][Y==j].astype('float32')) for i in range (X.shape[1])
for j in (np.unique(Y))])
Y_Data_reshaped = Y_Data.reshape(4, 3)
Y_Data_reshaped = np.swapaxes(Y_Data_reshaped, 0, 1)
X_axis = np.arange(len(columns)-1)
width = 0.25
# Plot the average
plt.bar(X_axis, Y_Data_reshaped[0], width, label = 'Setosa')
plt.bar(X_axis+width, Y_Data_reshaped[1], width, label = 'Versicolour')
plt.bar(X_axis+width*2, Y_Data_reshaped[2], width, label = 'Virginica')
plt.xticks(X_axis, columns[:4])
plt.xlabel("Features")
plt.ylabel("Value in cm.")
plt.legend(bbox_to_anchor=(1.3,1))
plt.show()
Step3: Split a dataset into training and testing datasets
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2)
Step4: Train the model
# Prediction of classifier svn = SVC() svn.fit(X_train, y_train)
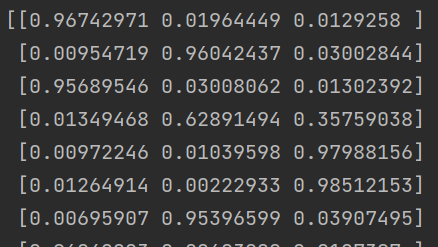
# Probability of each class svn_prob = SVC(probability=True) svn_prob.fit(X_train, y_train) pred_prob=svn_prob.predict_proba(X_test)
Step 5: Model Evaluation
pred = svn.predict(X_test)
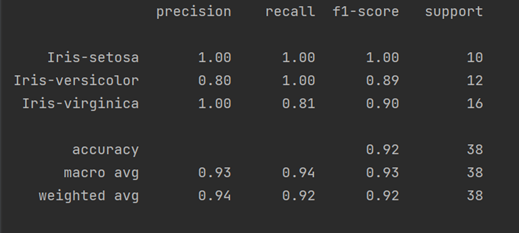
print("Accuracy Score:" + str(round(accuracy_score(y_test, pred),2)))

print(classification_report(y_test, pred))
Step 6: Testing the Model
X_new = np.array([[3, 2, 1, 0.2], [4.9, 2.2, 3.8, 1.1], [5.3, 2.5, 4.6, 1.9]])
pred_new = svn.predict(X_new)
print("Prediction of Species: {}".format(pred_new))

#Save the model
with open('SVM.pickle', 'wb') as f:
pickle.dump(svn, f)
# Load the model
with open('SVM.pickle', 'rb') as f:
model = pickle.load(f)
model.predict(X_new)