Project information
- Category: Machine Learning, Classification
- Source Data: Download
Project Details
Description
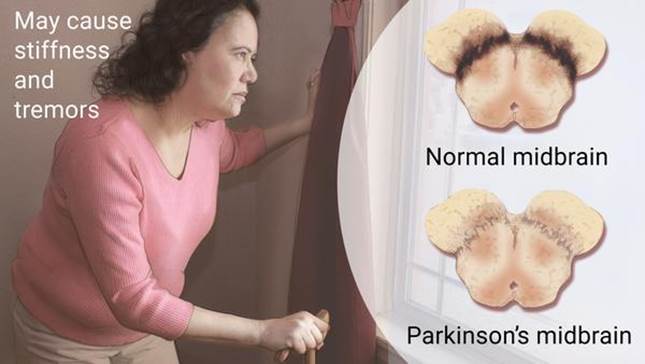
Parkinson's disease is a brain disorder that causes unintended or uncontrollable movements, such as shaking, stiffness, and difficulty with balance and coordination. Symptoms usually begin gradually and worsen over time. As the disease progresses, people may have difficulty walking and talking. The dataset contains two classes of status: 1 meaning yes and 0 meaning no and each class has 24 features.
Data Description:
The goal of the Parkinson’s disease is to train the random forest model, analyze importance of variables, tuning the accuracy of model, predict status based on their features.
As the dataset have labeled the variable, it is supervised machine learning. Supervised machine learning are types of machine learning that are trained on well-labeled training data. Labeled data means that training data is already tagged with correct output.
In this project, I will solve the problem using the algorithm: Random Forest as it is a supervised machine learning algorithm which analyzes data for classification and regression. Also, Random Forest Classification is one of the most robust classification methods.
How Random Forest Algorithms work?
Random forest algorithms have three main hyperparameters, which need to be set before training. These include node size, the number of trees, and the number of features sampled. From there, the random forest classifier can be used to solve for regression or classification problems.
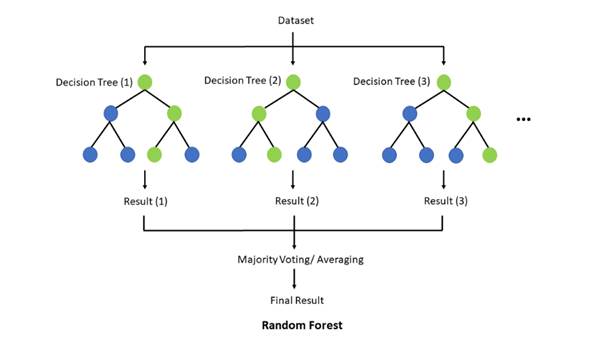
The random forest algorithm is made up of a collection of decision trees, and each tree in the ensemble is comprised of a data sample drawn from a training set with replacement, called the bootstrap sample. Of that training sample, one-third of it is set aside as test data, known as the out-of-bag (oob) sample, which we’ll come back to later. Another instance of randomness is then injected through feature bagging, adding more diversity to the dataset and reducing the correlation among decision trees. Depending on the type of problem, the determination of the prediction will vary. For a regression task, the individual decision trees will be averaged, and for a classification task, a majority vote—i.e. the most frequent categorical variable—will yield the predicted class. Finally, the oob sample is then used for cross-validation, finalizing that prediction.
Python Packages:
Roadmap:
Step 1:
Import packages:
#import libaries import pandas as pd import seaborn as sns import numpy as np from sklearn.model_selection import train_test_split from sklearn.metrics import accuracy_score from sklearn.ensemble import RandomForestClassifier from sklearn.ensemble import BaggingClassifier import matplotlib.pyplot as plt from sklearn.metrics import classification_report
url=r'url' df=pd.read_csv(url) print(df.head())
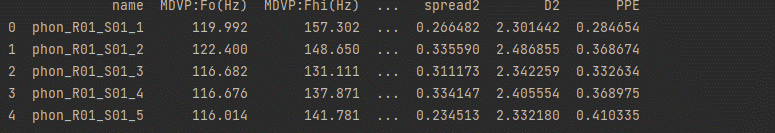
Step 2: Analyze and visualize the dataset
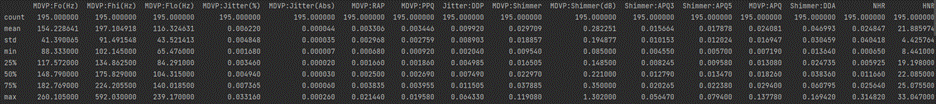
# display base statistical analysis regarding the data print(df.describe())
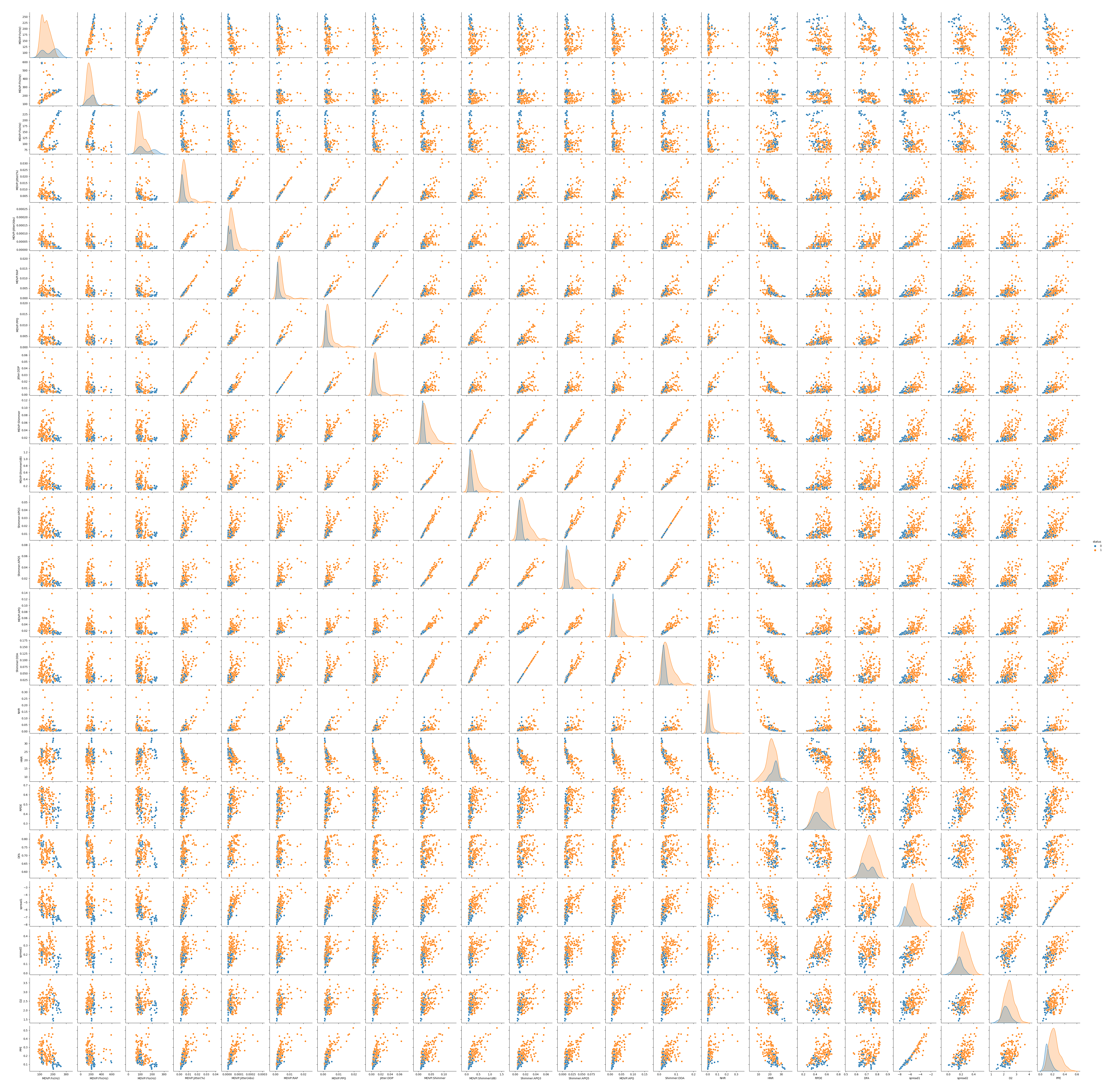
# visualize the dataset by labeled variable
sns.pairplot(df, hue='status')
plt.savefig('parkinsons variable analysis')
# Separate features and target
X=df.loc[:,df.columns!='status'].values[:,1:]
Y=df.loc[:,'status'].values
feature=df.drop('status', axis=1)
feature_list=list(feature.columns)
Y = data[:,4]
Step3: Split a dataset into training and testing datasets
# Split the dataset X_train,X_test,y_train,y_test=train_test_split(X, Y, test_size=0.2, random_state=42)
Step3: Train the model
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.20, random_state=42)
Step 4: Train the model
model=RandomForestClassifier(n_estimators=10,random_state=42) model.fit(X_train,y_train)
Step 5: Model Evaluation
pred=model.predict(X_test)
print('First Run Performance:'+str(accuracy_score(y_test,pred)*100))

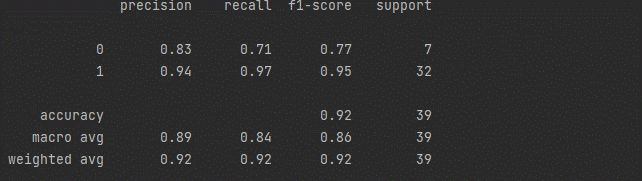
Step 6: Testing the Model
#Testing the model
X_new = np.array([[114.38000,130.10900,104.63400,0.00332,0.00003,0.00160,0.00199,0.00480,0.01503,0.13700,0.00812,0.00933,0.01133,0.02436,0.00401,26.00500,0.405991,0.761255,-5.966779,0.197938,1.974857,0.184067],
[117.38800,125.03800,110.97000,0.00346,0.00003,0.00169,0.00213,0.00507,0.01725,0.15500,0.00874,0.01021,0.01331,0.02623,0.00415,26.14300,0.361232,0.763242,-6.016891,0.109256,2.004719,0.174429],
[151.73700,190.20400,129.85900,0.00314,0.00002,0.00135,0.00152,0.00506,0.01469,0.13200,0.00728,0.00886,0.01230,0.02184,0.00570,24.15100,0.396610,0.745957,-6.486822,0.197919,2.449763,0.132703]])
pred_new = model.predict(X_new)
# expected results:0,0,1
print("Prediction of Species: {}".format(pred_new))

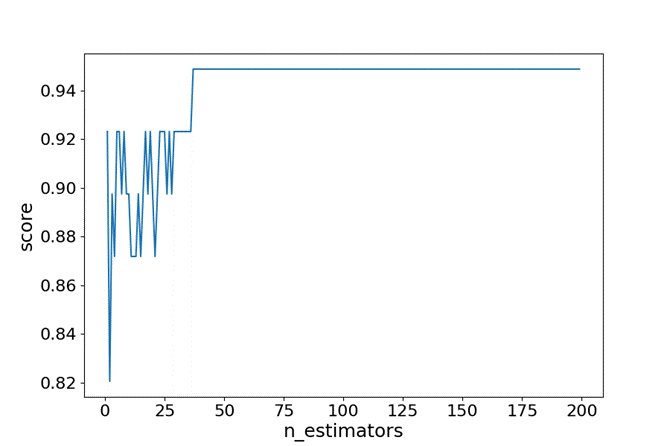
Step 7: Improving Performance
# bagging estimate
estimator_range = [e for e in range(1,200)]
models = []
scores = []
for n_estimators in estimator_range:
# Create bagging classifier
clf = BaggingClassifier(n_estimators = n_estimators, random_state = 42)
# Fit the model
clf.fit(X_train,y_train)
# Append the model and score to their respective list
models.append(clf)
scores.append(accuracy_score(y_true = y_test, y_pred = clf.predict(X_test)))
# Generate the plot of scores against number of estimators
plt.figure(figsize=(9,6))
plt.plot(estimator_range, scores)
# Adjust labels and font (to make visable)
plt.xlabel("n_estimators", fontsize = 18)
plt.ylabel("score", fontsize = 18)
plt.tick_params(labelsize = 16)
# Visualize plot
# plt.show()
plt.savefig('bagging estimation')
- Define the range from 1 to 200.
- Use bagging classifier to try one by one and log the result for later analysis.
- Plot the graph to assess the result and save it as output.
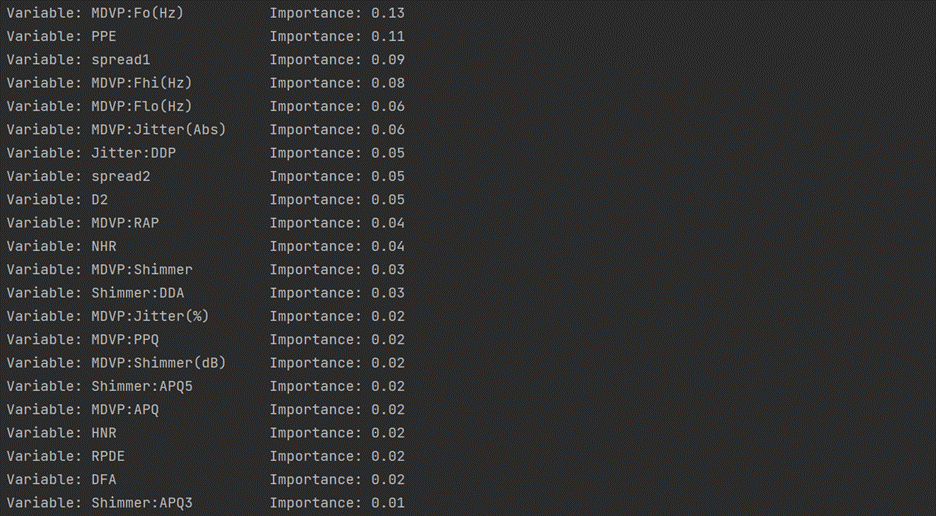
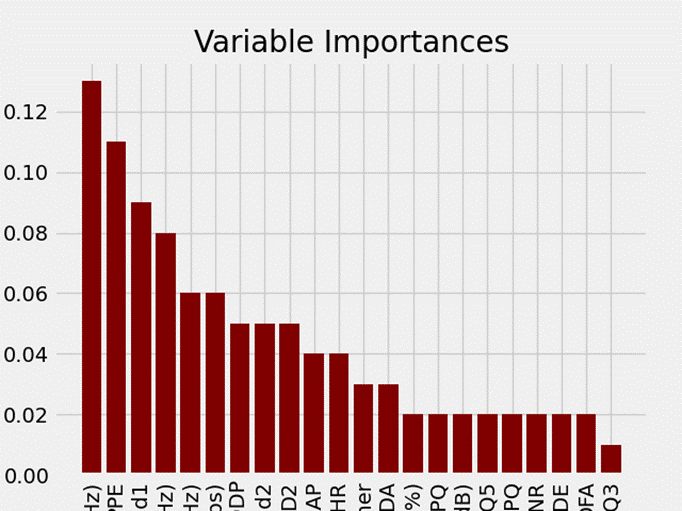
# Visualizations
# Make a bar chart
name=[]
value=[]
for pair in feature_importances:
name.append(pair[0])
value.append(pair[1])
plt.style.use('fivethirtyeight')
plt.bar(name, value, color='maroon', orientation = 'vertical')
# Tick labels for x axis
plt.xticks(rotation='vertical')
# Axis labels and title
plt.ylabel('Importance'); plt.xlabel('Variable'); plt.title('Variable Importances');
plt.show()
# Performence Improvement
# New random forest with only the two most important variables
rf_most_important = RandomForestClassifier(n_estimators=30,random_state=42)
# Extract the two most important features
important_indices=[feature_list.index('MDVP:Fo(Hz)'),feature_list.index('PPE'),feature_list.index('spread1'),feature_list.index('MDVP:Fhi(Hz)'),feature_list.index('MDVP:Flo(Hz)')
,feature_list.index('MDVP:Jitter(Abs)'),feature_list.index('Jitter:DDP'),feature_list.index('spread2'),feature_list.index('D2')]
X_train_important=X_train[:,important_indices]
X_test_important=X_test[:,important_indices]
# Train the random forest
rf_most_important.fit(X_train_important,y_train)
# Make predictions and determine the error
predictions=rf_most_important.predict(X_test_important)
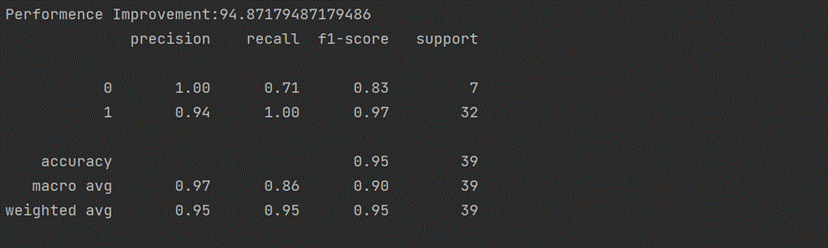
print('Performence Improvement:'+str(accuracy_score(y_test,predictions)*100))
print(classification_report(y_test, pred))