Project information
- Category: Modeling, Regression
- Source Data: statsmodels Package
Project Details
Data Description
Growth curves of pigs data contains weight of slaughter pigs measured weekly for 12 weeks. Data also contains the startweight (i.e. the weight at week 1). The treatments are 3 different levels of Evit = vitamin E (dose: 0, 100, 200 mg dl-alpha-tocopheryl acetat /kg feed) in combination with 3 different levels of Cu=copper (dose: 0, 35, 175 mg/kg feed) in the feed. The cumulated feed intake is also recorded. The pigs are littermates.
The goal of the Growth curves of pigs is to train mixed effect regression model, analyze the weight of pig, tuning the accuracy of model, predict weight based on their features.As the dataset has labeled the variable, it is supervised machine learning. Supervised machine learning is a type of machine learning that is trained on well-labeled training data. Labeled data means that training data is already tagged with correct output. In this project, I will solve the problem using the algorithm: mixed effect regression as data is longitudinal. Also, mixed effect regression is one of the most robust regression methods.
What is mixed effect regression?
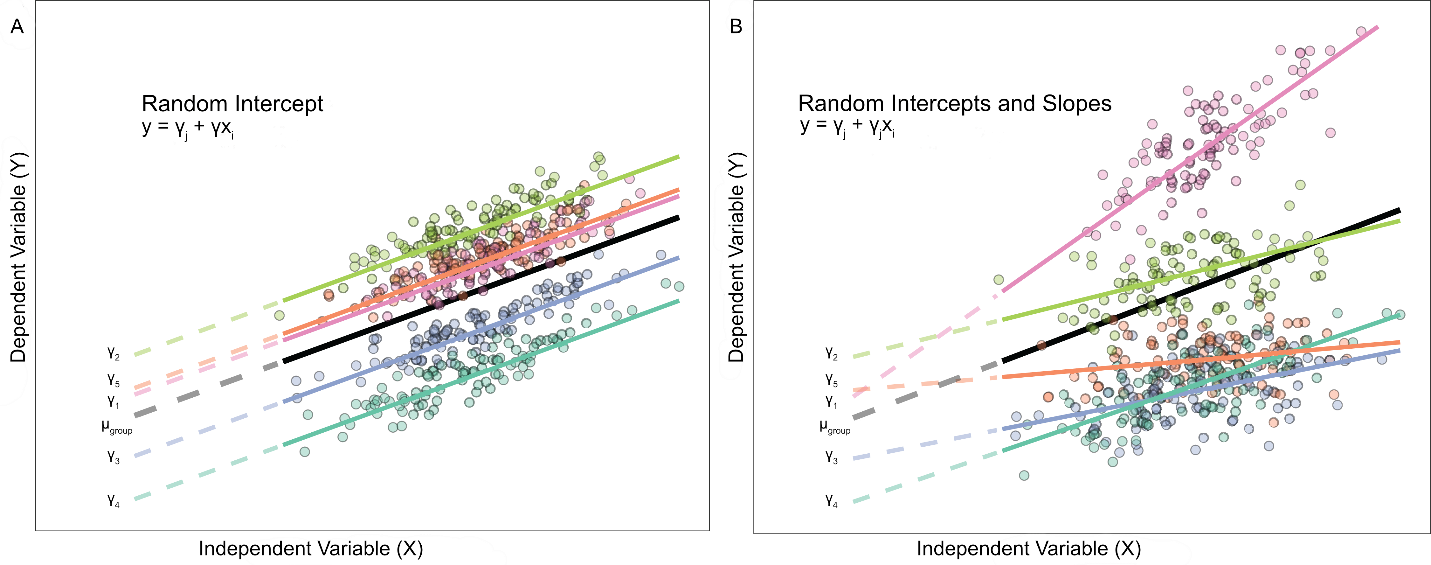
Mixed effects regression is an extension of the general linear model (GLM) that considers the hierarchical structure of the data. Mixed effect models are also known as multilevel models, hierarchical models, mixed models (or specifically linear mixed models (LMM)) and are appropriate for many types of data such as clustered data, repeated-measures data, longitudinal data, as well as combinations of those three. Since mixed effects regression is an extension of the general linear model, let's recall that the general linear regression equation looks like:

Fixed factors are the independent variables that are of interest to the study, e.g. treatment category, sex or gender, categorical variable, etc.
Random factors are the classification variables that the unit of anlaysis is grouped under, i.e. these are typically the variables that define level 2, level 3, or level n. These factors can also be thought of as a population where the unit measures were randomly sampled from, i.e. (using the school example from above) the school (level 2) had a random sample of students (level 1) scores selected to be used in the study.
Python Packages:
Roadmap:
Step 1:
Import packages:
#import libaries import statsmodels.api as sm import statsmodels.formula.api as smf import researchpy as rp import matplotlib.pyplot as plt import scipy.stats as stats import seaborn as sns from statsmodels.stats.diagnostic import het_white import warnings
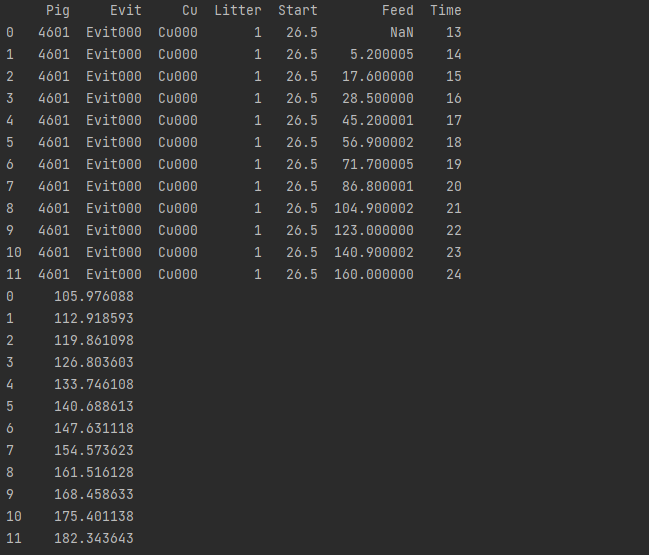
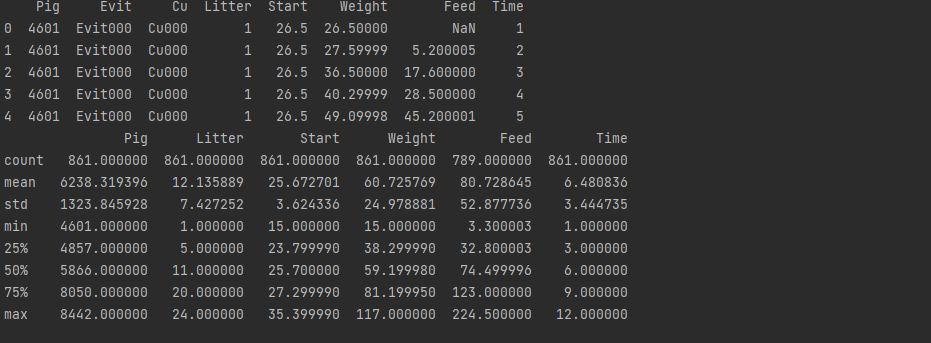
df = sm.datasets.get_rdataset('dietox', 'geepack').data
print(df.head())
print(df.describe())
 <
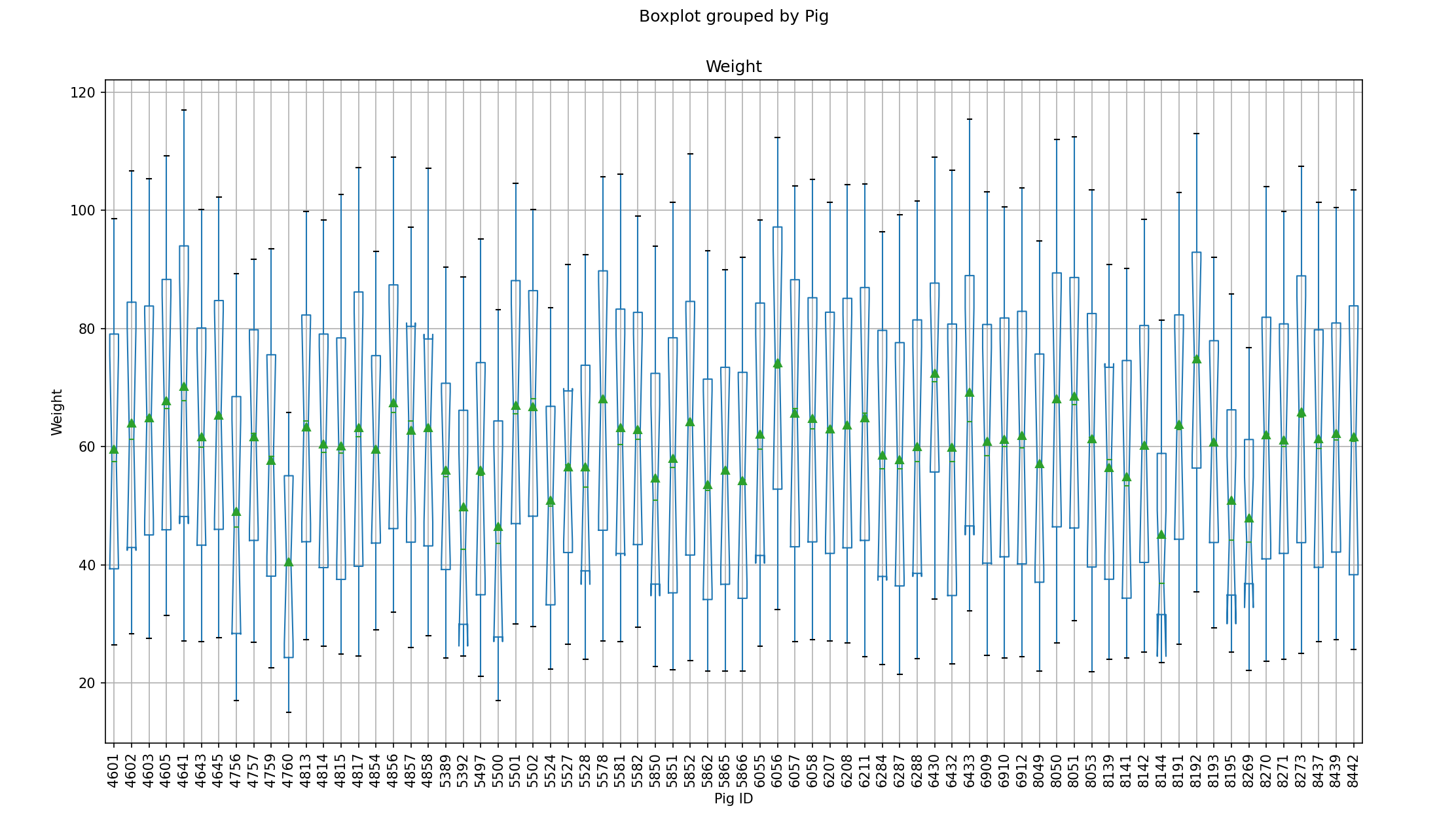
<Step2: Analyze and visualize the dataset
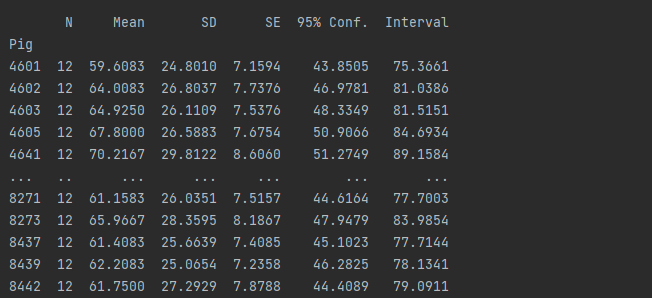
print(rp.summary_cont(df.groupby(['Pig'])['Weight']))
boxplot = df.boxplot(["Weight"], by = ["Pig"],
figsize = (16, 9),
showmeans = True,
notch = True)
boxplot.set_xlabel("Pig ID")
boxplot.set_ylabel("Weight")
plt.xticks(rotation='vertical')
plt.show()
Step 3: Train the model
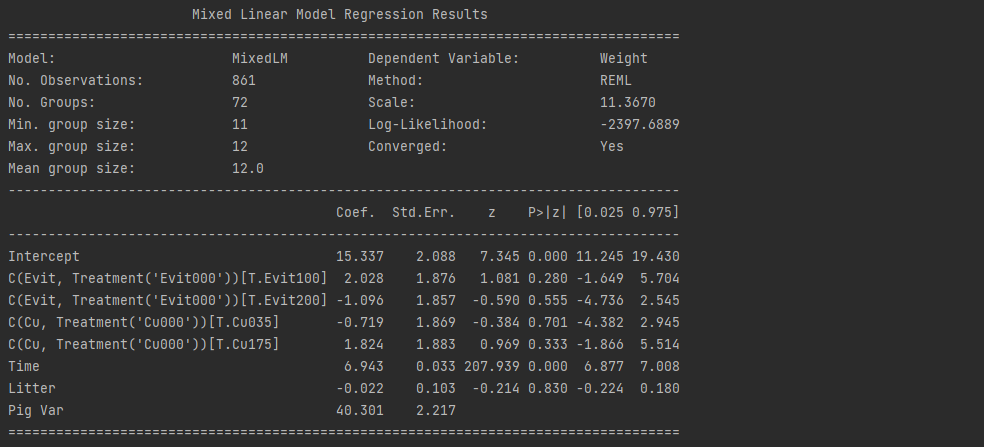
# Random intercepts
model1 = smf.mixedlm("Weight ~ Time+C(Evit,Treatment('Evit000'))+C(Cu,Treatment('Cu000'))+Litter",df,groups= "Pig").fit()
print("Random intercepts")
print(model1.summary())
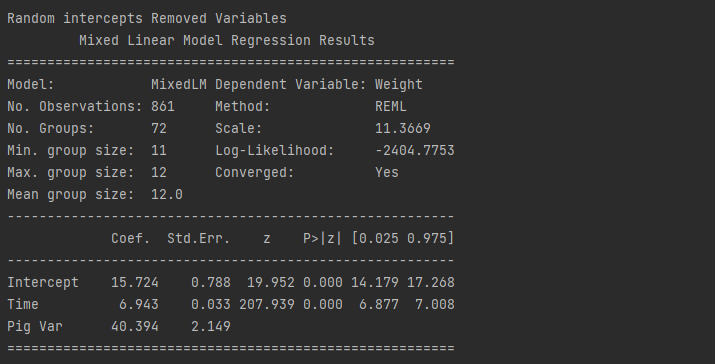
# Random intercepts removed variables
model11 = smf.mixedlm("Weight ~ Time",df,groups= "Pig").fit()
print("Random intercepts Removed Variables")
print(model11.summary())
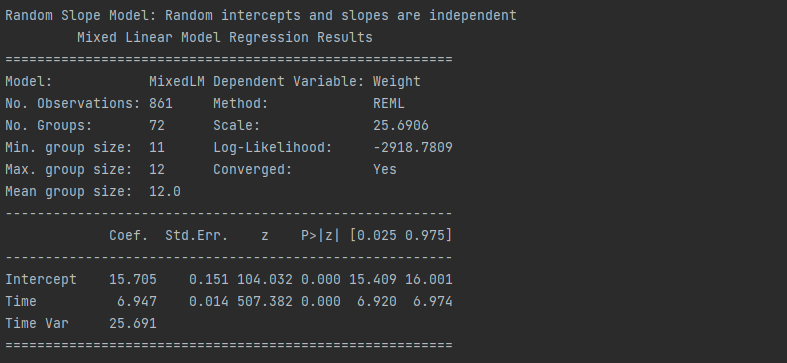
# Random Slope Model: Random intercepts and slopes are independent
model2 = smf.mixedlm("Weight ~ Time",df,groups= "Pig", vc_formula = {"Time" : "0 + C(Time)"}).fit()
print("Random Slope Model: Random intercepts and slopes are independent")
print(model2.summary())
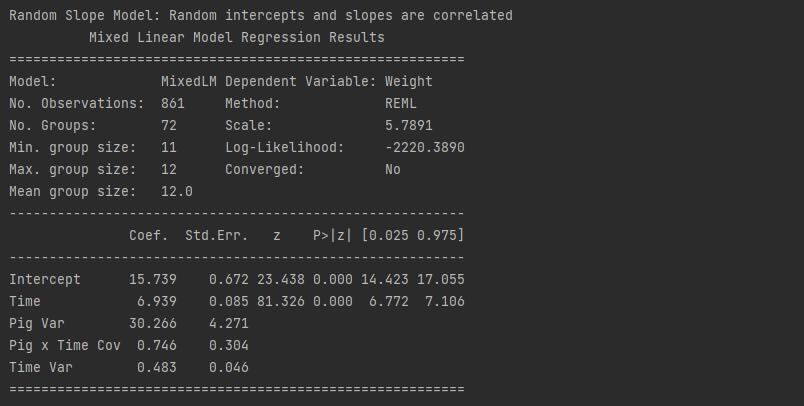
# Random Slope Model: Random intercepts and slopes are correlated
model3 = smf.mixedlm("Weight ~ Time",df,groups= "Pig", re_formula="Time").fit()
print("Random Slope Model: Random intercepts and slopes are correlated")
print(model3.summary())
Step 4: assumption check
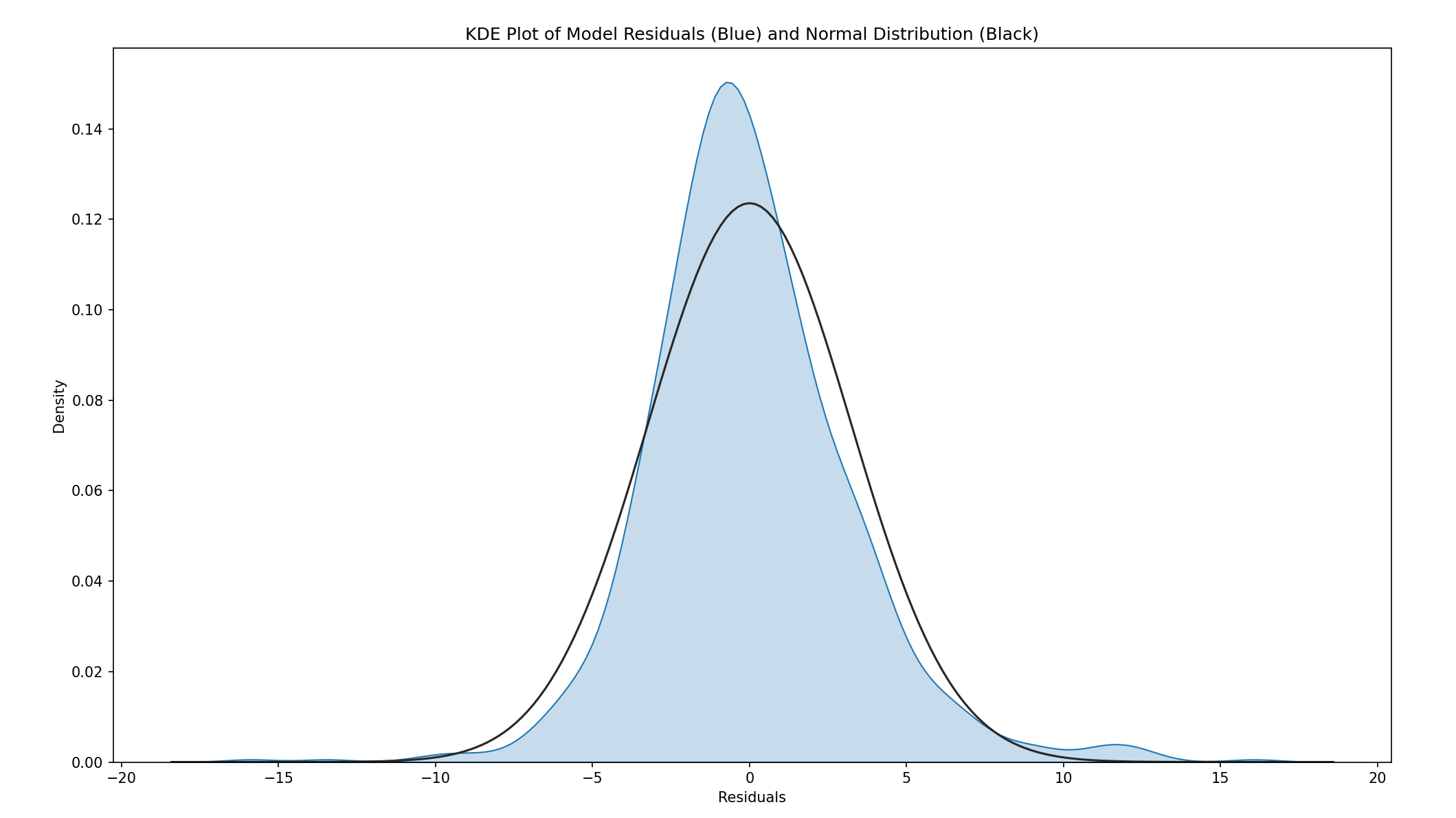
#NORMALITY
fig = plt.figure(figsize = (16, 9))
ax = sns.distplot(model3.resid, hist = False, kde_kws = {"shade" : True, "lw": 1}, fit = stats.norm)
ax.set_title("KDE Plot of Model Residuals (Blue) and Normal Distribution (Black)")
ax.set_xlabel("Residuals")
plt.show()
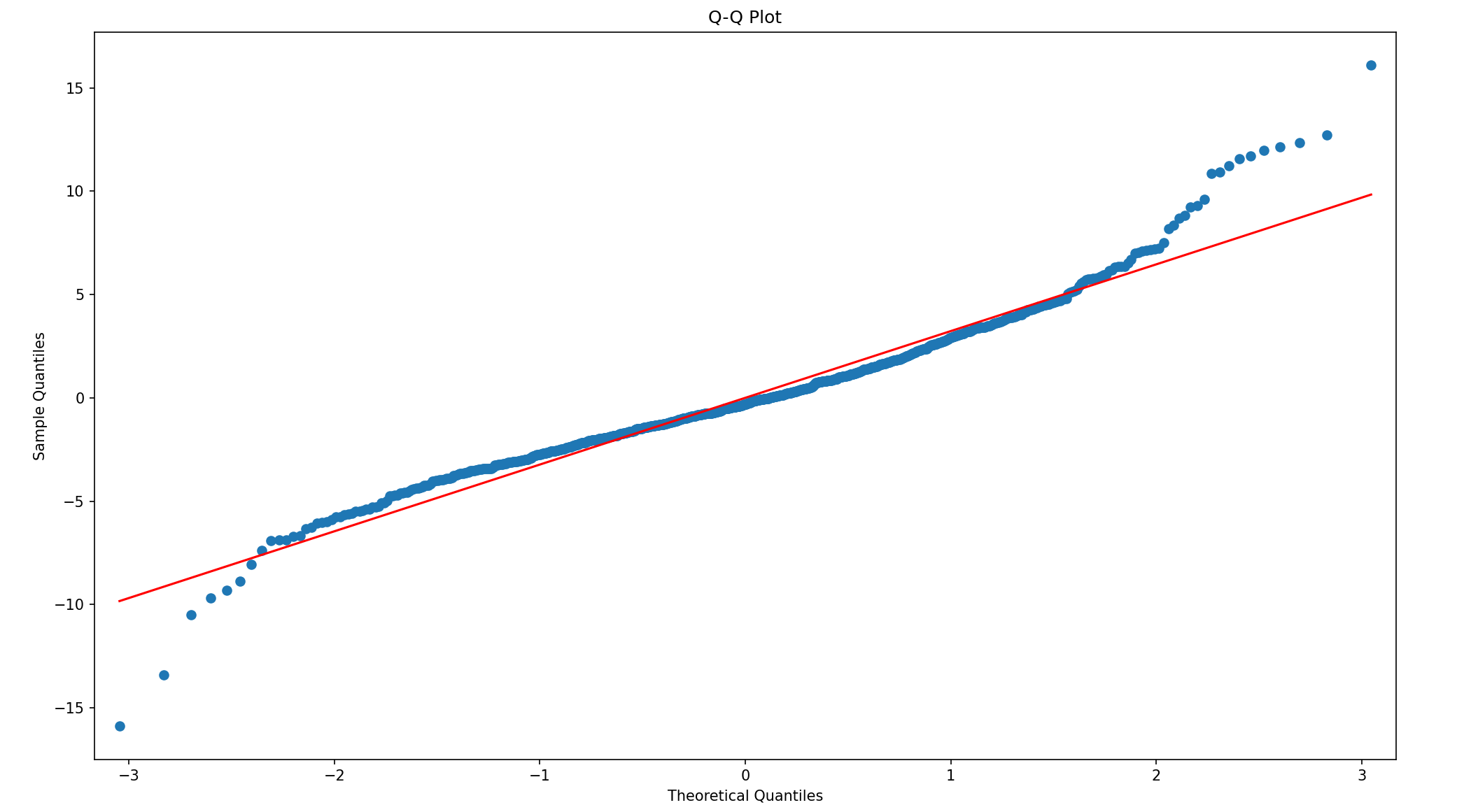
## Q-Q PLot
fig = plt.figure(figsize = (16, 9))
ax = fig.add_subplot(111)
sm.qqplot(model11.resid, dist = stats.norm, line = 's', ax = ax)
ax.set_title("Q-Q Plot")
plt.show()
# Shapir-Wilk test of normality
labels = ["Statistic", "p-value"]
norm_res = stats.shapiro(model11.resid)
for key, val in dict(zip(labels, norm_res)).items():
print(key, val)

# homoskedasticity
fig = plt.figure(figsize = (16, 9))
ax = sns.scatterplot(y = model11.resid, x = model11.fittedvalues)
ax.set_title("RVF Plot")
ax.set_xlabel("Fitted Values")
ax.set_ylabel("Residuals")
plt.show()
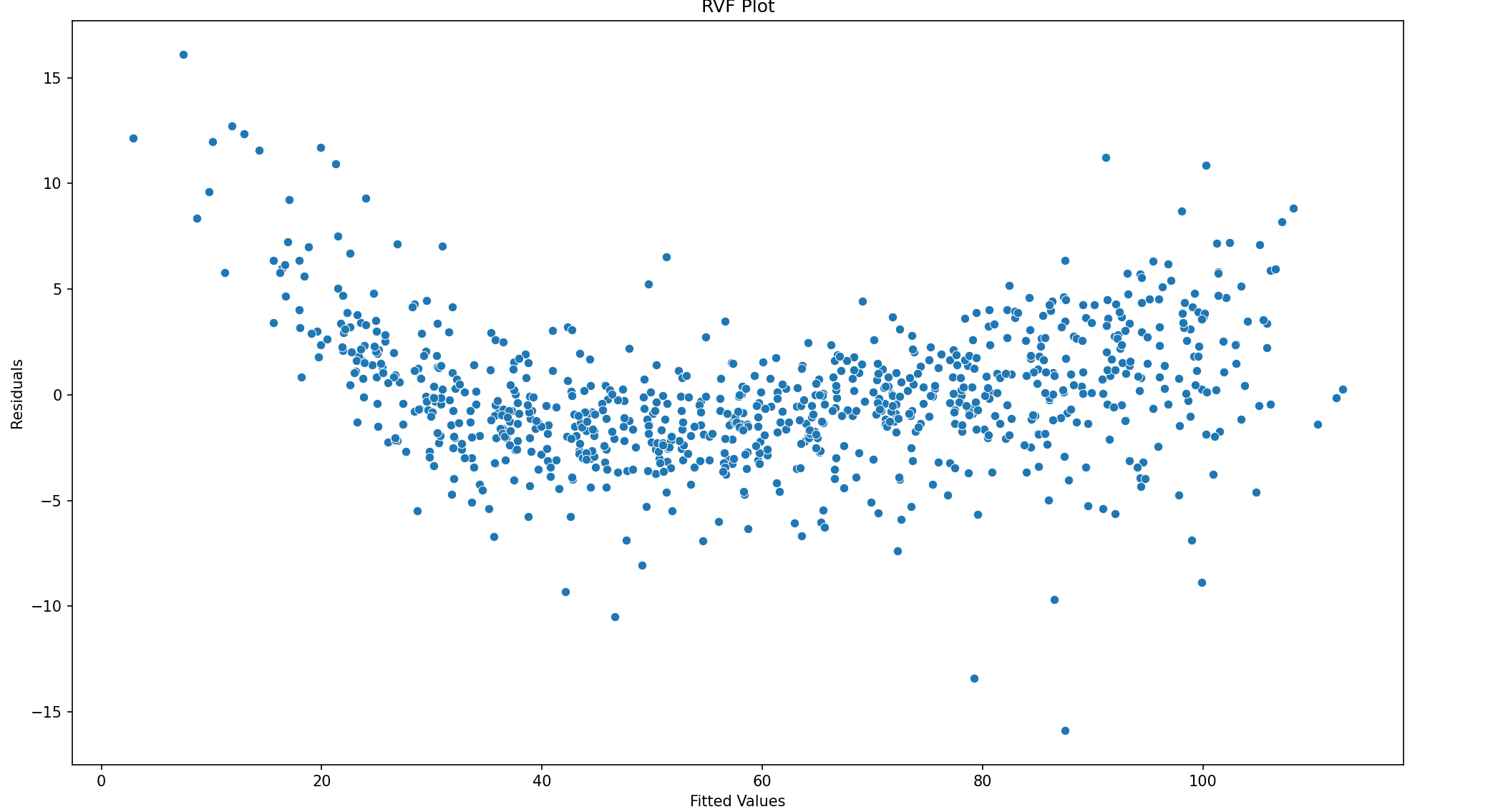
fig = plt.figure(figsize = (16, 9))
ax = sns.boxplot(x = model11.model.groups, y = model11.resid)
ax.set_title("Distribution of Residuals for Weight by Pig ID")
ax.set_ylabel("Residuals")
ax.set_xlabel("Pig ID")
plt.xticks(rotation='vertical')
plt.show()

Step 5: test the model
# Testing the model test_file_path=r'C:\Users\shang\Desktop\ML\data\pig_test.csv' test=pd.read_csv(test_file_path) print(test) pred=model11.predict(exog=test) print(pred)